Research: Evolution of Germline Development
Nearly all sexually-reproducing animals produce germ cells (cells that give rise to eggs and sperm). Germ cells are totipotent - they can produce all cell types in organisms. Proper specification of germ cells is essential for life; failure to make germ cells can lead to sterility. To preserve their totipotency, germ cells must be protected from factors that would cause them to differentiate into somatic cells (such as muscle, neurons, skin, etc.).
Germ cells in multiple organisms have common traits, including the expression of a highly-conserved DEAD-box RNA helicase Vasa, the presence of specific chromatin methylation patterns, and cessation of transcription. However, organisms appear to use a variety of mechanisms to ensure that germ cells acquire the correct fate at the correct time. How diverse are these mechanisms? Are there common rules that govern germline development? To better understand how germ cell fate is specified, and how germline development has evolved among animals, I am studying this question in two invertebrate species.
Tardigrades as a new model system for germline development
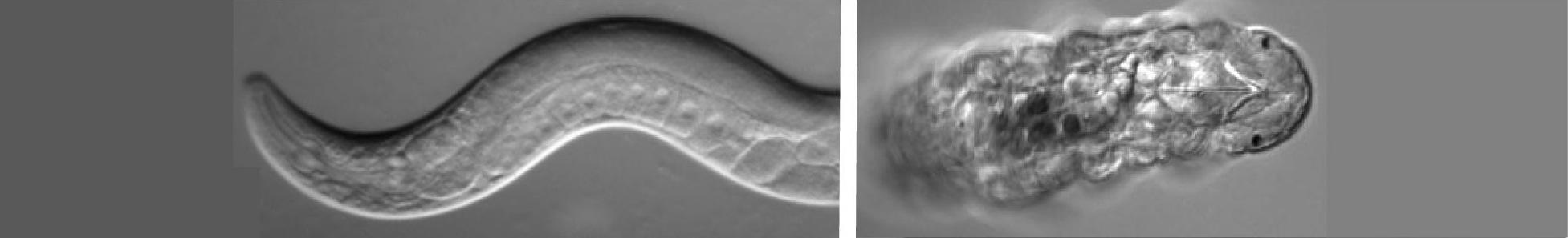
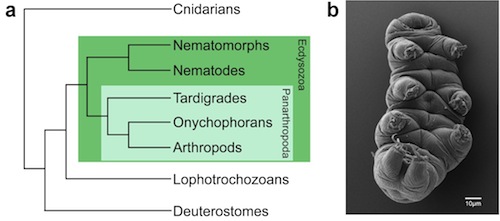 I have been developing the tardigrade Hypsibius dujardini as a model for understanding germ cell fate specification. Tardigrades (also known as water bears) are small, segmented invertebrates that have become popular organisms for studying survival in extreme conditions. Tardigrades comprise a phylum closely related to arthropods, nematodes and other Ecdysozoans (invertebrates that molt) (Fig. 1), and their evolutionary position can help us to better understand how similarities and differences in germline development have arisen in well-studied model organisms.
I have been developing the tardigrade Hypsibius dujardini as a model for understanding germ cell fate specification. Tardigrades (also known as water bears) are small, segmented invertebrates that have become popular organisms for studying survival in extreme conditions. Tardigrades comprise a phylum closely related to arthropods, nematodes and other Ecdysozoans (invertebrates that molt) (Fig. 1), and their evolutionary position can help us to better understand how similarities and differences in germline development have arisen in well-studied model organisms.
In 2013, I published a protocol to disrupt gene function by RNA interference (RNAi). RNAi is a well-conserved mechanism by which messenger RNA encoding a known gene is degraded, preventing translation of the encoded protein, and producing "loss-of-function" phenotypes (Fig. 2). In 2015, the H. dujardini draft genome sequence was published, and is now available for searching through NCBI. With this data, we can identify putative homologs of candidate germline genes in H. dujardini, and disrupt their function by RNAi.
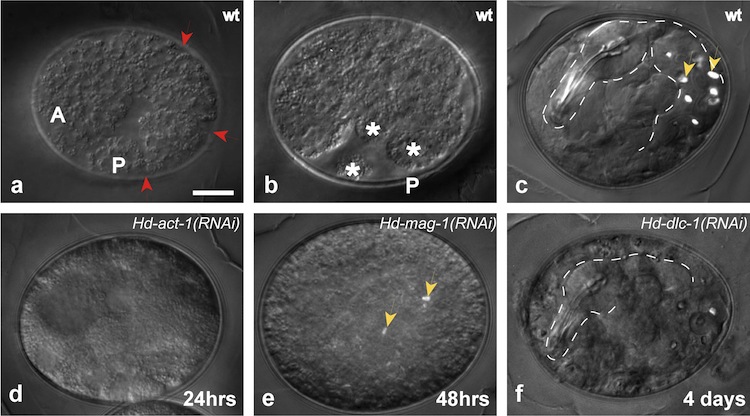
Fig. 2: RNAi results in target-specific depletion of gene function. a-c Representative images of wild-type embryos. a Stage 13 embryo (~24 hours after egg-laying), showing elongation along the anterior-posterior axis, with ectodermal segmentation (red arrows). b Same embryo as in (a), at late Stage 15 (~48 hrs after egg-laying), showing three developing limb buds (asterisks). Intestinal birefringent granules are visible in a higher focal plane. c Stage 19 embryo (~4 days after egg laying), prior to hatching. The pharynx and intestine are outlined (white dotted lines). Yellow arrows in this panel and in panel (e) mark birefringent granules, a marker of intestine differentiation. Note that this embryo is different from the one shown in a, b. d Hd-act-1(RNAi) embryo, ~24 hours after egg-laying. e Hd-mag-1(RNAi) embryo, ~48 hrs after egg-laying. The embryo has not elongated along the anterior-posterior axis, but birefringent granules are visible. f Hd-dlc-1(RNAi) embryo, ~4 days after egg-laying. The pharynx and part of the intestine are outlined (white dotted lines). The intestine appears to lack birefringent granules. A=anterior; P=posterior.
Some of the projects that I hope to have SPU students working on include:
- Analysis of tardigrade ultrastructure by transmission electron microscopy: germ cells in many organisms contain electron-dense granules that are visible by TEM. We do not yet know if germ cells in H. dujardini possess these electron-dense granules.
- Characterization of H. dujardini germ cells: germ cells in many organisms express highly-conserved proteins required for germ cell specification and/or maintenance. In this project, students would use antibodies specific to these proteins to ask which proteins are expressed in H. dujardini germ cells, and when this protein expression is first detected during development.
- Cloning and characterization of DEAD-box RNA helicases: these proteins, homologs of the highly-conserved germline protein Vasa, are required for germline development in many organisms. This project would clone different Vasa homologs from H. dujardini, and use RNAi to ask what roles these genes may play in development.
Germline development in nematodes
As another approach to understanding the evolution of germline development, I am interested in the extent to which mechanisms of germline development are conserved within the nematode phylum. Much of what we know about cell fate specification and early germline development in nematodes comes from studies in C. elegans. However, it is not clear how cell fates are specified in other nematode species. Analysis of multiple nematode genome sequences suggests that some germline-specific proteins required for C. elegans development are absent in non-Caenorhabditis species. What mechanisms do non-Caenorhabditis species use to ensure the proper segregation of somatic and germ cell fates?
Some of the projects that I hope to have SPU students working on (both in my lab, and in lab courses that I teach) include:
- Microscopic analysis of germline development: using live microscopy, film embryo development, and try to identify general patterns of development that can be used as a benchmark to characterize loss-of-function mutant embryos.
- Cloning and characterization of genes required for germ cell specification: using available gene sequence databases for different nematode species, we will identify C. elegans genes required for germline development, and use RNAi to determine if these genes are also required for germline development in those species.
- Identification of novel factors required for germline development: using RNAi and/or mutagenesis screens, we will identify genes that when mutated, prevent germline development in other nematodes.
Note: I am open to mentoring other projects if students have specific questions they are interested in studying using tardigrades or nematodes, and that can be studied using the resources available in my lab and at SPU.
Research: Genetic diversity of an island deer population
Since 2015, I have collaborated with Dr. Eric Long, SPU Professor of Biology and black-tailed deer specialist, on two projects related to the population of black-tailed deer that live on Blakely Island (in the San Juan archipelago). The overall goal of these projects is to better understand the effects of geographic isolation on these deer, which have no natural predators and are the largest non-human mammals on the island.
- Genetic diversity of Blakely Island deer: using DNA extracted from deer tissue, we are amplifying and sequencing genetic markers from mitochondrial and genomic DNA. Analysis of these sequences will reveal the number of unique variants (alleles) of a particular marker that are present in the population, and provide insight into the diversity of the deer population.
- Analysis of juvenile mortality: deer skulls have been collected from juveniles that died of natural causes, but which died before obvious signs of sexual dimorphism (such as the presence of antlers in male deer). We are establishing protocols to extract and amplify DNA sequences from deer skulls that will allow us to determine the sex of each deer.